Laboratory of Structure-Function Based Drug Design
|
|
CONTACTS |
|
|
|
|
|
|
|
|
TEACHING BY RESEARCH
THE MAIN DIRECTIONS OF RESEARCH AND DEVELOPMENT
Based on the application of existing and development of new computer methods using machine learning and artificial intelligence:
– Exploration and extraction from the texts of scientific publications and other sources data and knowledge about biological activity and metabolism of drug-like chemical compounds, genes, proteins and their relationships with physiological and pathological processes in the human body.
– Analysis of the “structure-activity”, “structure-property” and “structure-structure” relationships of drug-like chemical compounds, assessment of their biological activity spectra and estimation of safety, taking into account the relationships of pharmacological effects and molecular mechanisms of action and the network of biotransformations in biological entities.
– Investigations of the structural and functional features of biological macromolecules, identification of functionally significant fragments involved in protein-ligand interactions, as well as prediction of drug resistance based on the analysis of nucleotide/amino acid sequences, taking into account the individual characteristics of the patient.
– Modeling of regulatory processes in signaling networks to identify the promising pharmacological targets and their combinations and predict the molecular mechanisms of the manifestation of the main pharmacological and adverse effects of drugs in the human body.
– Design and development of the Way2Drug web portal, which provides in silico studies of the “Disease – Target - Biological Process - Pharmacological Action” relationships based on analysis of big data by the systems pharmacology methods.
The staff of Laboratory provides lectures and seminars on Bioinformatics and Computer-Aided Drug Discovery at the Medical-Biological Faculty of N.I. Pirogov Russian National Research Medical University and other Universities in Moscow, and for IBMC Ph.D. Students in specialty 1.5.8 - Mathematical Biology and Bioinformatics.
Students from several Moscow Universities (N.I. Pirogov Russian National Research Medical University, M.V. Lomonosov Moscow State University, Sechenov University, etc.) perform their practical and diploma studies in the Laboratory. The best students continue their postgraduate studies, to earn Ph.D, Degree in specialty 1.5.8 - Mathematical Biology and Bioinformatics or specialty 1.5.4 - Biochemistry.
Corresponding Member of the Russian Academy of Sciences, Professor Vladimir Poroikov regularly presents lectures for scientists, young researchers, postgraduate and graduate students within the framework of the Skolkovo Open University “Pharma's Cool” (2015, 2019), Kazan Summer Schools in Chemoinformatics (2013, 2015, 2017, 2019), Marina Astvatsaturyan Author’s Program for Physicians at the First Medical Channel “Medicine in Context” (2019), XIX V.A. Nasonova Russian School of Rheumatologists “Rheumatology: Systemic Nature” (2020), XIII V.I. Pokrovsky Annual Russian Congress on Infectious Diseases (2021), International Scientific and Practical Conference «Scientific, Technological and Innovative Cooperation of the BRICS Countries» (2022), V International Scientific Forum «Step into the Future: Artificial Intelligence and Digital Economy. Technoeconomic: Transformation of Platforms» (2023), within the framework of the project «Technological Sovereignty. Natural Choice?» answers to questions from students of non-medical universities are presented (2023), etc.
Professor Oleg Gomazkov published a number of conceptual monographs about the brain (Brain Aging and Neurotrophic Therapy, 2011; Neurogenesis as an Adaptive Function of the Adult Brain, 2013); Astrocytes - Stars That Control the Brain, 2018); about the COVID-19 pandemic (Endothelium is the Target Chosen by the Coronavirus, 2020; Pathogenesis of Vascular Lesions, or the Devil Is in the Details, 2021; Four Stories on the Topic of COVID, 2023); and others, on the basis of which lectures are regularly presented for physicians and young scientists in Russia and abroad.
Since 1995, Annual Symposia “Bioinformatics and Computer-Aided Drug Discovery” have been held; since 2021, they have become International and are implemented as virtual conferences. The Jubilee, XXX Symposium on Bioinformatics and Computer-Aided Drug Discovery will take place in 2024. Within the framework of these Symposia, Young Scientists’ Contests are held; the winners are awarded by diplomas.
DEVELOPMENT OF BIO- AND CHEMOINFORMATICS METHODS AND THEIR APPLICATIONS
In 2021-2023, along with the studies performed in the framework of the Program of Basic Scientific Research in the Russian Federation for the long-term period (2021-2030) and projects supported by the Russian Science Foundation, we carried out investigations under contracts with Russian Pharmaceutical Companies (OJSC "Avexima", LLC "Pharmasoft", LLC "AlexAnn", LLC "PeptidPro", as well as the State Institute of Medicines and Good Practices of the Ministry of Industry and Trade of Russia. Direct involvement in the research and development carried out by domestic pharma companies allows us to better understand the real needs of the industry and adapt our computer-aided drug design methods to effectively fulfill the appropriate tasks.
A web portal has been implemented to provide informational-analytical support to participants in domestic projects to search for anti-coronavirus drugs, which includes informational and computational components. Informational component provides information about: (i) COVID-19 disease; (ii) biological processes affected by SARS-CoV-2 coronavirus infection; (iii) molecular targets, which inhibiting allows to block pathological processes; (iv) the possibilities of drugs repositioning for the treatment of COVID-19, etc. The computational component is represented by five web applications: (i) Repositioning - search by structural similarity among more than four thousand pharmaceutical substances approved for medical use; (ii) Anti-Covid-19 allows one to predict 4 types of anti-coronavirus activity based on the structural formula of the analyzed compound; (iii) Comorbidity allows you to evaluate the biological activity profile of the analyzed compound in relation to the possibility of treating 57 concomitant SARs-CoV-2 diseases using a specialized version of PASS; (iv) Multi-targeting allows one to evaluate the overall profile of biological activity, which includes 1957 of the most important effects and mechanisms of action, using PASS Refined; (v) Toxicity allows for the assessment of side effects and toxicity for the analyzed compounds using a specialized version of PASS.
In silico screening was carried out among 1.08 billion structures for 4 SARS-CoV-2 targets (3CLpro, PLpro, RdRp, TMPRSS2) within the framework of the European Initiative “Grand Challenge Against COVID-19”. We selected 10 thousand “hits”, and we were included in 20 out of 130 groups whose proposals were selected for experimental testing. 36 of the proposed molecules have been synthesized; for one molecule the predicted activity (PLpro inhibition) has been confirmed experimentally. Based on in silico assessments, we also selected a number of drugs (Narlaprevir, Boceprevir, Disulfiram, etc.), which anti-coronavirus activity was later shown in in vitro experiments. However, the project organizers refused to synthesize and test the activity of the repositioned drugs, despite the initial conditions for participation required to perform the virtual screening among drugs approved for medical use.
A set of methods, algorithms, computer programs and web resources has been created for analyzing the biological activity profiles of drug-like compounds, taking into account their metabolism in the human body based on the generation of metabolic networks and predicting the biological activity spectra of drug-like organic compounds (the study was supported by the Russian Science Foundation Grant No. 19-15-00396).
A web platform “hiv-host” for analyzing interactions between HIV and the human body has been developed, including: (i) RHIVDB database containing information about the medications taken by a patient with a specific prevailing variant of HIV and the amino acid sequences of the main proteins for this variant; (ii) HIV-host database on the interaction between HIV and the human body; (iii) Web service for predicting the efficacy of antiretroviral therapy and a model for predicting the rate of progression of HIV infection; (iv) HIV-synergy web service for predicting drug combinations synergism in antiretroviral activity.
A method has been developed for extracting the names of drug-like compounds slowing the progression of HIV infection and the new coronavirus infection from the texts of scientific publications. This method was used to assess the mechanisms of progression of HIV infection and new coronavirus infection. (the study was supported by the Russian Science Foundation Grant No. 19-75-10097).
An approach has been developed to analyze changes in gene expression for patients with different rates of progression of HIV infection. The mechanisms mediated different rates of development of HIV infection in the human body have been established and key regulators of these changes have been identified. An approach has been developed to analyze the efficacy of antiretroviral therapy based on individual patient characteristics, including age, gender, and gene expression profile. The method was tested in a prospective clinical study; changes in the expression level were determined for genes selected on the basis of their potential contribution to the progression of HIV infection, and taking into account their significance for the efficacy of antiretroviral therapy (the study was supported by the Russian Science Foundation Grant No. 19-75-10097).
The models have been developed for in silico prediction of drug-drug interactions (DDI). They are freely available on the Internet as the DDI-Pred service. Using this service, one can use the structural formulae of a pair of drug-like substances to obtain estimates of various types severity of drug-drug interactions, as well as associated adverse drug reactions from the cardiovascular system (the study was supported by the Russian Science Foundation Grant No. 17-75-20250).
Machine learning methods have been developed to assess whether HIV variants belong to the class of variants that are resistant or sensitive to antiretroviral therapy. It has been shown that the average prediction accuracy for various drugs is 81-93%. The results obtained can be used to optimize therapy for patients with HIV/AIDS (the studies was supported by the Russian Foundation for Basic Research Grant No. 16-34-60187 and Russian Science Foundation Grant No. 17-75-20250).
Based on in silico analysis of the effects of phytocomponents of medicinal plants of traditional Indian medicine and network pharmacology methods, several new potential pharmacological targets for the treatment of vascular dementia were identified. The effectiveness of inhibiting the identified targets was confirmed in experimental animal models of the disease (the study was supported by the Russian Foundation for Basic Research - DST Grant No. 16 -54-45016).
A virtual screening of compounds potentially active against M. Tuberculosis was carried out among 200 thousand molecules from the repository of the N.D. Zelinsky Institute of Organic Chemistry of the Russian Academy of Sciences. 30 selected compounds were tested in the REMA (REsazurin Microtiter Assay) test system in vitro against M. tuberculosis (strain H37Rv) at the Federal Research Center “Fundamental Principles of Biotechnology” of the Russian Academy of Sciences. 12 active compounds from various chemical classes were identified with MICs ranging from 2.2 – 16.7 μM.
An original method of analysis of amino acid sequences based on local similarity has been developed, which has essential advantages in performance and flexibility in comparison with commonly used approaches based on the alignment of the primary structure of proteins. Computer programs for the analysis of amino acid sequences have been implemented that allow establishing the structure-function relationships and identifying functionally essential sites. (Supported by the RFBR grants No. 16-04-00491 and 19-015-00374)
An innovative system of atom-centric molecular descriptors has been proposed, which allows characterizing intermolecular interactions and, on this basis, analyzing various structure-property relationships. These are MNA (Multilevel Neighborhoods of Atoms), QNA (Quantitative Neighborhoods of Atoms), LMNA (Labeled Multilevel Neighborhoods of Atoms), etc.)
A novel method has been implemented for analyzing quantitative structure-activity relationships (QSAR) and predicting the activity of new substances; its advantages have been demonstrated in comparison with the number of other widely used QSAR methods. (Supported by the European grants No. FP6 LSHB-CT-2007-037590 и FP7 grant 200787)
Using PASS Online, anti-inflammatory effects were predicted in 13 dithioloquinolinethione derivatives that had previously been shown to have antimicrobial effects. Experimental studies on a carrageenan-induced inflammation model confirmed the presence of activity comparable in magnitude to the activity of indomethacin. A structural similarity search of the Cortellis Drug Discovery Intelligence database did not reveal structural analogues with anti-inflammatory activity. Thus, based on the PASS prediction, a new class of substances with anti-inflammatory activity has been discovered.
An original fragment-based drug design method is proposed for the design of physiologically active substances with the required properties using molecular descriptors and algorithms developed in our laboratory. Using this approach, anti-inflammatory drugs with dual mechanisms of action (cyclooxygenase 1, 2 and lipoxygenase inhibitors) were designed, synthesized, and tested experimentally.
A novel method is proposed for in silico generating combinatorial libraries of macrolides with a simultaneous assessment of the properties of the generated molecules, to select the substances with desired properties, following the further design and preparation of the corresponding producer strains by genetic engineering methods. (Supported by the IFTI grant No. RU.55229907.00160)
A technique for dichotomic modelling of processes in regulatory signalling networks has been developed to identify promising molecular targets, the inhibition of which leads to a blockage of the cell cycle or the switching of tumour cells into apoptosis. Its validation on the case study of breast cancer allowed identifying pharmacological targets and their combinations and, based on virtual screening of over 24 million chemical compounds from the ChemNavigator library, to reveal promising new inhibitors that exhibit synergism with the RITA substance, the well-known P53 protein reactivator suppressed in many types of tumours. The activity of the compounds found has been confirmed by in vivo in experiments on xenograft mice with transfected human tumors. (Supported by the European grant No. FP6 LSHB-CT-2007-037590)
The possibilities of predicting resistance to antiretroviral drugs based on the analysis of amino acid and nucleotide sequences to optimize HIV/AIDS therapy have been investigated. Based on the analysis of nucleotide sequences of HIV-1 protease and reverse transcriptase, we studied the possibilities to predict the effectiveness of antiretroviral drugs combinations to optimize the HIV/AIDS treatment. . (Supported by the RSCF grants No. 17-75-10187 and 19-75-10097).
An approach is proposed to determine the mechanisms of adverse drugs effects at different levels of biological organization, based on the assessment of the profiles of the effects of drug substances on human proteins; analysis of drug-induced changes in gene expression with the search for genes whose expression change correlates with an adverse effect; examining the role of identified proteins and genes in known signalling pathways to establish those pathways for which exposure plays a crucial role in inducing a side effect. The developed approach was validated on the example of evaluating the mechanisms of side effects of drugs on the cardiovascular system and hepatobiliary system. (Supported by the RSCF grant No. 17-75-10168).
A method has been developed for the integrated in silico assessment of the toxicity of xenobiotics, taking into account their metabolism in the human body, allowing to evaluate the metabolic pathways of compounds in the human body and to evaluate their acute, specific (cardiotoxicity, hepatotoxicity, nephrotoxicity) and chronic (carcinogenicity, teratogenicity, mutagenicity, the effect on the reproductive system) toxicity. (Supported by the RSCF grant No. 14-15-00449).
The analysis of existing approaches to computer-aided prediction of multitarget action and pleiotropic effects of pharmaceutical substances has been performed. It was concluded that with the growth of chemogenomic data and expansion of the studied chemical space, the multitarget QSAR modelling would become more common than single-target QSAR studies and that multitarget QSAR will lead to the discovery of novel medicines with much-improved safety and potency profiles. (Supported by the Russian State Academies of Sciences Fundamental Research Program for 2013-2020).
Using our computer-aided drug design methods, new physiologically active compounds have been found with nootropic, anxiolytic, antidepressant, anticonvulsant, antidiabetic, antitumor, antimicrobial, antifungal, antiretroviral and several other types of biological activity.
A previously unknown nootropic effect is predicted in antihypertensive drugs – angiotensin-converting enzyme inhibitors (perindopril, quinapril, etc.), confirmed by the experiment and, subsequently, in the clinical trials.
Based on a computer-aided estimation, anti-inflammatory action of the antibacterial drug clarithromycin was predicted and confirmed by the experiment, due to which this drug can be used for the treatment of Inflammatory Bowel Disease. (Supported by the European grant No. 305564).
Using our computational methods, new piperazine derivatives, TRPC6 (Transient Receptor Potential Canonical 6) channel agonists, have been discovered. They may be further developed as potential pharmacological substances for the treatment of Alzheimer’s disease.
An analysis of the hidden pharmacological potential of the phytocomponents from some preparations of traditional Indian Ayurveda medicine is carried out. New types of biological activity predicted for several natural substances have been experimentally confirmed. (Supported by the RFBR grant No. 16-54-45016).
Using the proprietary technology for the analysis of large chemical data, we analyzed 961 million structures of easily synthesized compounds from the SAVI library (Synthetically Accessible Virtual Inventory). Potential inhibitors of HIV-1 protease (53 compounds) and reverse transcriptase (48 compounds), as well as agonists of Toll-like 7 (53 compounds) and Toll-like 8 (1378 compounds) receptors and STING (627 compounds), affecting the innate immunity. Information about these molecules has been passed on to our partners at National Cancer Institute (NIH) for the synthesis and biological testing; for the four Toll-like receptors of subtype 7 selected by us, the predictions are confirmed experimentally; the other experimental studies continues. (Supported by the RFBR grant No. 17-54-30015-NIH_a).
Based on our computational methods, it was found that the metabolism of the original Russian drug phenazepam is carried out with the participation of CYP3A4; the in silico prediction is confirmed by in vitro and in vivo experiments. (Supported by the RSCF grant No. 14-15-00449).
It was predicted that the metabolism of the anticoagulant drug phenindione is carried out with the participation of cytochrome P450 1A2, rather than 2C9, as previously thought, which allowed us to explain the absence of cytochrome gene polymorphism CYP2C9 influence on the action of the drug in the clinic, rationalize the drug treatment of atrial fibrillation options and increase patient adherence therapy by reducing hemorrhagic complications. (Supported by the RSCF grant No. 14-15-00449).
An in silico analysis of the possible drug-drug interaction of umifenovir (arbidol), studied in the clinic as a potential pharmaceutical agent for COVID-19 therapy, was carried out. The results indicate a low probability of umifenovir interaction with paracetamol and dabigatran. However, for the anticoagulants of acenocoumarol and warfarin, the maximum severity of possible interaction with umifenovir (class 1 according to the ORCA classification) is predicted, which may be due to the inhibition of CYP2C9. (Supported by the RSCF grant No. 17-75-20250).
The hypothesis about the role of astroglial cells associated with component neurons in providing neurotransmitter function of the brain is substantiated. Data analysis showed that astrocytes are integral elements of information neural networks that affect synaptic processes due to modulation of neurotransmitter activity. Due to the interaction of gliotransmitters with neurons, astrocytes are involved in the regulation of memory and complex behavioural processes in normal and pathological states of the brain.
- Olga Bocharova, Blokhin National Medical Research Center of Oncology, Moscow, Russia.
- Valery Dembitsky, Centre for Applied Research and Innovation, Lethbridge College, Lethbridge, Canada.
- James Devillers, Centre for the Treatment of the Scientific Information (CTIS), Rillieux La Pape, France.
- Tamara Dzagurova, M.P. Chumakov Federal Scientific Center for Research and Development of Immunobiological Drugs of the Russian Academy of Sciences (Polio Institute), Moscow, Russia.
- Alexey Egorov, M.V. Lomonosov Moscow State University, Moscow, Russia.
- Athina Geronikaki, Aristotle University of Thessaloniki, Thessaloniki, Greece.
- Rajesh Goel, Panjabi University, Patiala, India.
- Marina Gottikh, A.N. Belozersky Institute of Physico-Chemical Biology MSU, Moscow, Russia.
- Alan Kaluev, Sirius University, Krasnodar Region, Federal Territory "Sirius", Russia.
- Alexander Kel, GeneXplain GmbH, Germany.
- Natasha Kirienko, Rice University, Houston, TX, USA.
- Alexander Knyazev, Lobachevsky University, Nizhny Novgorod, Russia.
- Fedor Kolpakov, Sirius University, Krasnodar Region, Federal Territory "Sirius", Russia.
- Alexander Kryukov, Department of Clinical Pharmacology, O.M. Filatov Hospital Number 15, Moscow, Russia.
- Lyubov Kozlovskaya, M.P. Chumakov Federal Scientific Center for Research and Development of Immunobiological Drugs of the Russian Academy of Sciences (Polio Institute), Moscow, Russia.
- Vladimir Luzhanin, Perm Chemical-Pharmaceutical Academy, Perm, Russia.
- Irina Lupanova, Russian Research Institute of Medicinal and Aromatic Plants (VILAR), Moscow, Russia.
- José L. Medina-Franco, National Autonomous University of Mexico, Mexico.
- Irina Milentieva, Kemerovo State University, Kemerovo, Russia.
- Marc Nicklaus, National Cancer Institute, National Institute of Health. NCI-Frederick, MD, USA.
- Kyoung Tai No, Yonsei University, Seoul, Republic of Korea.
- Dmitry Osolodkin, M.P. Chumakov Federal Scientific Center for Research and Development of Immunobiological Drugs of the Russian Academy of Sciences (Polio Institute), Moscow, Russia.
- Maria Povydysh, Saint Petersburg Chemical-Pharmaceutical University, Saint Petersburg.
- Alexander Raikov, Jinan Institute of Supercomputing Technology, Shandong, China
- Kunal Roy, Jadavpur University, Jadavpur, India.
- Elena Salina, Federal Research Center “Fundamentals of Biotechnology” of the Russian Academy of Sciences, Moscow, Russia.
- Narahari G. Sastry, CSIR Indian Institute of Chemical Technologies, Hyderabad, India.
- Marcus Tullius Scotti, Universidade Federal da Paraíba, Brazil.
- Viktor Semenov, N.D. Zelinsky Institute of Organic Chemistry, Moscow, Russia.
- Dmitry Sychev, Russian Medical Academy of Continuous Professional Education, Moscow, Russia.
- Khidmet Shikhaliev, Voronezh State University, Voronezh, Russia.
- Andrey Shchekhotikhin, Gause Institute of New Antibiotics, Moscow, Russsia.
- Alexander Tropsha, School of Pharmacy, University of North Carolina at Chapel Hill, USA.
- Alexandre Varnek, Louis Pasteur University (Strasbourg), France.
- Pavel Vassil’ev, Volgograd State Medical University, Volgograd, Russia.
- Tatiana Voronina, V.V. Zakusov Research Institute of Pharmacology, Moscow, Russia.
- Walter Filgueira de Azevedo Junior, Federal University of Alfenas, Alfenas, Brazil.
- Weiliang Zhu, Shanghai Institute of Materia Medica, Chinese Academy of Sciences, Shanghai, China.
- Roman Zubarev, Karolinska Institutet, Stokholm, Sweden.
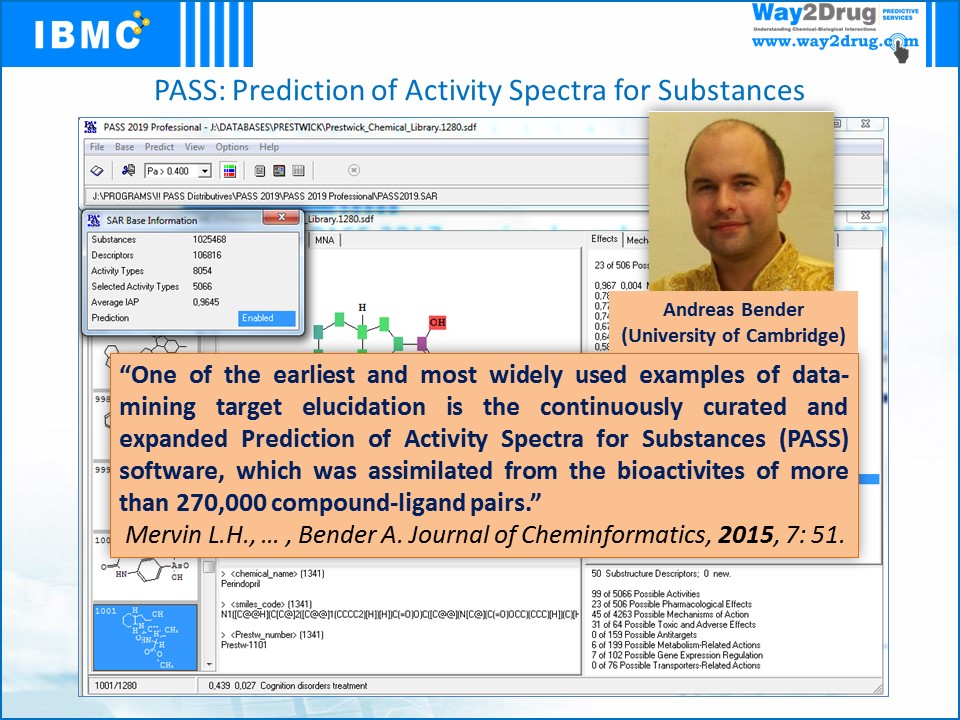
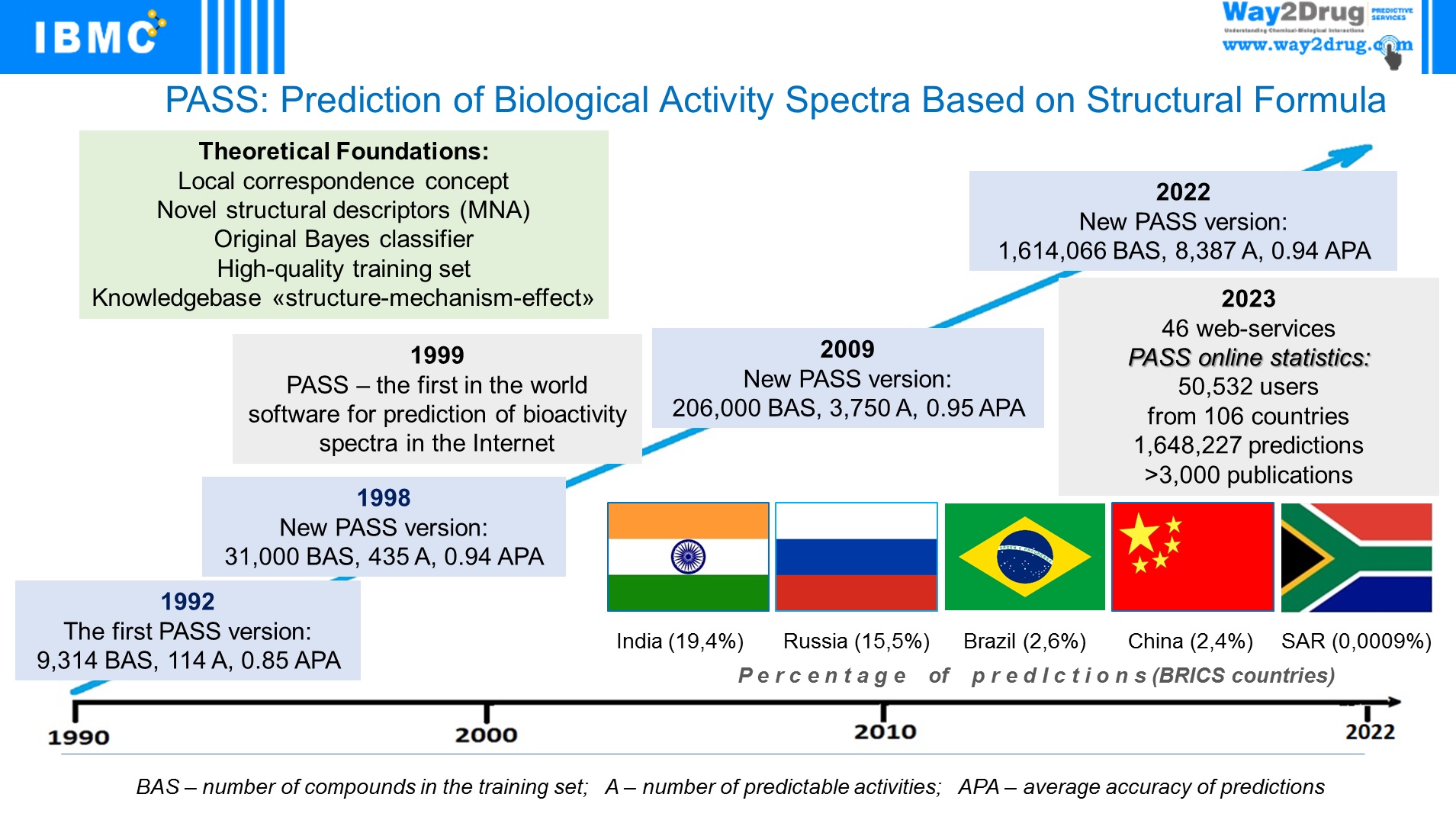
The first in the world freely available Internet resource PASS Online has been implemented, predicting over 8000 types of biological activity with an average accuracy of about 93%.
The Way2Drug informational-computational platform has been created, on the basis of which over forty freely accessible web resources are presented. Links to some of these web resources are provided below:
PASS – Prediction of Activity Spectra for Substances:
- PASS Online, prediction of 4000 types of biological activity.
- Possibility to select specialized versions of PASS, including PASS 2022, and predictions using one or more versions of the program.
Selected areas of pharmaceutical agents’ action:
- PASS Targets, prediction of interaction with molecular targets.
- KinScreen, prediction of interaction of pharmacological substances with human kinome.
- CLC Pred 2.0 - prediction of cytotoxicity against tumor and non-tumor cell lines.
- DIGEP-Pred, prediction of drug-induced gene expression.
Side effects and toxicity:
- ADVER-Pred, prediction of adverse drugs effects on cardiovascular and hepatobiliary systems.
- hERG-Pred, prediction of hERG-channels inhibition.
- ROSC-Pred, prediction of organ-specific carcinogenicity.
- Acute Rat Toxicity, prediction of rats acute toxicity using four routes of administration of a pharmacological substance.
- Antitarget Prediction, prediction of interaction with undesirable targets.
- DDI-Pred, prediction of drug-drug interactions.
Metabolism of pharmacological substances and their permeability through the BBB:
- Metabolic Stability - prediction of metabolic stability of pharmaceutical substances.
- SOMP, prediction of metabolism sites for pharmacological agents.
- RA, prediction of reacting atoms during the biotransformation.
- SMP, prediction of substrate’s and metabolite’s specificity to the biotransformation enzymes.
- MetaTox, prediction of toxicity taking into account the metabolism of drug.
- MetaPASS, prediction of biological activity profiles of the parent substance and its metabolites.
- BBB Permeability - prediction of permeability through the blood-brain barrier (BBB).
Web-platform for analyzing interactions between HIV and the human body:
- RHIVDB database - information about medications taken by a patient with a specific prevalent HIV variant and the amino acid sequences of the main structural proteins for this variant.
- HIV-host database – information about the interaction of HIV and the human body.
- TR - web service for predicting the efficacy of antiretroviral therapy and models for predicting the rate of progression of HIV infection.
- AntiHIV-synergy - web service for predicting drug combinations antiretroviral activity synergy.
Currently, our web services are used by over 50 thousand scientists, postgraduate, graduate and undergraduate students from 106 countries to select the most promising molecules for synthesis and determine the optimal directions for testing their biological activity.
Databases
- WWAD (World-Wide Approved Drugs) – database of pharmaceutical substances approved for medicinal use in 52 countries.
- Phyto4Health - database on phytocomponents of medicinal plants included in Russian Pharmacopoeia.
- HGMMX - database on the metabolism of xenobiotics by human microbiome.
![]() Schimunek J., Seidl P., Elez K., Hempel T., Le T., Noé F., Olsson S., Raich L., Winter R., Gokcan H., Gusev F., Gutkin E.M., Isayev O., Kurnikova M.G., Narangoda C.H., Zubatyuk R., Bosko I.P., Furs K.V., Karpenko A.D., Kornoushenko Y.V., Shuldau M., Yushkevich A., Benabderrahmane M.B., Bousquet-Melou P., Bureau R., Charton B., Cirou B.C., Gil G., Allen W.J., Sirimulla S., Watowich S., Antonopoulos N.A., Epitropakis N.E., Krasoulis A.K., Pitsikalis V.P., Theodorakis S.T., Kozlovskii I., Maliutin A., Medvedev A., Popov P., Zaretckii M., Eghbal-Zadeh H., Halmich C., Hochreiter S., Mayr A., Ruch P., Widrich M,. Berenger F., Kumar A., Yamanishi Y., Zhang K.Y.J., Bengio E., Bengio Y., Jain M.J., Korablyov M., Liu C.H., Marcou G., Glaab E., Barnsley K., Iyengar S.M., Ondrechen M.J., Haupt V.J., Kaiser F., Schroeder M., Pugliese L., Albani S.., Athanasiou C., Beccari A., Carloni P., D`Arrigo G., Gianquinto E., Goßen J., Hanke A., Joseph B.P., Kokh D.B., Kovachka S., Manelfi C., Mukherjee G., Muñiz-Chicharro A., Musiani F., Nunes-Alves A., Paiardi G., Rossetti G., Sadiq S.K., Spyrakis F., Talarico C., Tsengenes A., Wade R.C., Copeland C., Gaiser J., Olson D.R., Roy A., Venkatraman V., Wheeler T.J., Arthanari H., Blaschitz K., Cespugli M., Durmaz V., Fackeldey K., Fischer P.D., Gorgulla C., Gruber C., Gruber K., Hetmann M., Kinney J.E., Padmanabha Das K.M., Pandita S., Singh A., Steinkellner G., Tesseyre G., Wagner G., Wang Z.F., Yust .RJ., Druzhilovskiy D.S., Filimonov D.A., Pogodin P.V., Poroikov V., Rudik A.V., Stolbov L.A., Veselovsky A.V., De Rosa M., De Simone G., Gulotta M.R., Lombino J., Mekni N., Perricone U., Casini A., Embree A., Gordon D.B., Lei D., Pratt K., Voigt C.A., Chen K.Y., Jacob Y., Krischuns T., Lafaye P., Zettor A., Rodríguez M.L., White K.M., Fearon D., Von Delft F., Walsh M.A., Horvath D., Brooks C.L. 3rd, Falsafi B., Ford B., García-Sastre A., Yup Lee S., Naffakh N., Varnek A., Klambauer G., Hermans T.M. (2024) A community effort in SARS-CoV-2 drug discovery, Molecular Informatics, 43(1), e202300262. DOI:10.1002/minf.202300262
Schimunek J., Seidl P., Elez K., Hempel T., Le T., Noé F., Olsson S., Raich L., Winter R., Gokcan H., Gusev F., Gutkin E.M., Isayev O., Kurnikova M.G., Narangoda C.H., Zubatyuk R., Bosko I.P., Furs K.V., Karpenko A.D., Kornoushenko Y.V., Shuldau M., Yushkevich A., Benabderrahmane M.B., Bousquet-Melou P., Bureau R., Charton B., Cirou B.C., Gil G., Allen W.J., Sirimulla S., Watowich S., Antonopoulos N.A., Epitropakis N.E., Krasoulis A.K., Pitsikalis V.P., Theodorakis S.T., Kozlovskii I., Maliutin A., Medvedev A., Popov P., Zaretckii M., Eghbal-Zadeh H., Halmich C., Hochreiter S., Mayr A., Ruch P., Widrich M,. Berenger F., Kumar A., Yamanishi Y., Zhang K.Y.J., Bengio E., Bengio Y., Jain M.J., Korablyov M., Liu C.H., Marcou G., Glaab E., Barnsley K., Iyengar S.M., Ondrechen M.J., Haupt V.J., Kaiser F., Schroeder M., Pugliese L., Albani S.., Athanasiou C., Beccari A., Carloni P., D`Arrigo G., Gianquinto E., Goßen J., Hanke A., Joseph B.P., Kokh D.B., Kovachka S., Manelfi C., Mukherjee G., Muñiz-Chicharro A., Musiani F., Nunes-Alves A., Paiardi G., Rossetti G., Sadiq S.K., Spyrakis F., Talarico C., Tsengenes A., Wade R.C., Copeland C., Gaiser J., Olson D.R., Roy A., Venkatraman V., Wheeler T.J., Arthanari H., Blaschitz K., Cespugli M., Durmaz V., Fackeldey K., Fischer P.D., Gorgulla C., Gruber C., Gruber K., Hetmann M., Kinney J.E., Padmanabha Das K.M., Pandita S., Singh A., Steinkellner G., Tesseyre G., Wagner G., Wang Z.F., Yust .RJ., Druzhilovskiy D.S., Filimonov D.A., Pogodin P.V., Poroikov V., Rudik A.V., Stolbov L.A., Veselovsky A.V., De Rosa M., De Simone G., Gulotta M.R., Lombino J., Mekni N., Perricone U., Casini A., Embree A., Gordon D.B., Lei D., Pratt K., Voigt C.A., Chen K.Y., Jacob Y., Krischuns T., Lafaye P., Zettor A., Rodríguez M.L., White K.M., Fearon D., Von Delft F., Walsh M.A., Horvath D., Brooks C.L. 3rd, Falsafi B., Ford B., García-Sastre A., Yup Lee S., Naffakh N., Varnek A., Klambauer G., Hermans T.M. (2024) A community effort in SARS-CoV-2 drug discovery, Molecular Informatics, 43(1), e202300262. DOI:10.1002/minf.202300262
![]() Pogodin P.V., Salina E.G., Semenov V.V., Raihstat M.M., Druzhilovskiy D.S., Filimonov D.A., Poroikov V.V. (2024) Ligand-based virtual screening and biological evaluation of inhibitors of Mycobacterium tuberculosis H37Rv, SAR and QSAR in Environmental Research, 35(1), 53-69. DOI:10.1080/1062936X.2024.2304803
Pogodin P.V., Salina E.G., Semenov V.V., Raihstat M.M., Druzhilovskiy D.S., Filimonov D.A., Poroikov V.V. (2024) Ligand-based virtual screening and biological evaluation of inhibitors of Mycobacterium tuberculosis H37Rv, SAR and QSAR in Environmental Research, 35(1), 53-69. DOI:10.1080/1062936X.2024.2304803
![]() Lagunin A.A., Rudik A.V., Pogodin P.V., Savosina P.I., Tarasova O.A., Dmitriev A.V., Ivanov S.M., Biziukova N.Y., Druzhilovskiy D.S., Filimonov D.A., Poroikov V.V. (2023) CLC-Pred 2.0: A Freely Available Web Application for In Silico Prediction of Human Cell Line Cytotoxicity and Molecular Mechanisms of Action for Druglike Compounds, International Journal of Molecular Sciences, 24(2), 1689. DOI:10.3390/ijms24021689
Lagunin A.A., Rudik A.V., Pogodin P.V., Savosina P.I., Tarasova O.A., Dmitriev A.V., Ivanov S.M., Biziukova N.Y., Druzhilovskiy D.S., Filimonov D.A., Poroikov V.V. (2023) CLC-Pred 2.0: A Freely Available Web Application for In Silico Prediction of Human Cell Line Cytotoxicity and Molecular Mechanisms of Action for Druglike Compounds, International Journal of Molecular Sciences, 24(2), 1689. DOI:10.3390/ijms24021689
![]() Ionov N., Druzhilovskiy D., Filimonov D., Poroikov V. (2023) Phyto4Health: Database of Phytocomponents from Russian Pharmacopoeia Plants, Journal of Chemical Information and Modeling, 63(7), 1847-1851. DOI:10.1021/acs.jcim.2c01567
Ionov N., Druzhilovskiy D., Filimonov D., Poroikov V. (2023) Phyto4Health: Database of Phytocomponents from Russian Pharmacopoeia Plants, Journal of Chemical Information and Modeling, 63(7), 1847-1851. DOI:10.1021/acs.jcim.2c01567
![]() Ivanov S.M., Tarasova O.A., Poroikov V.V. (2023) Transcriptome-based analysis of human peripheral blood reveals regulators of immune response in different viral infections, Frontiers in Immunology, 14, 1199482. DOI:10.3389/fimmu.2023.1199482
Ivanov S.M., Tarasova O.A., Poroikov V.V. (2023) Transcriptome-based analysis of human peripheral blood reveals regulators of immune response in different viral infections, Frontiers in Immunology, 14, 1199482. DOI:10.3389/fimmu.2023.1199482
![]() Kolodnitsky A.S., Ionov N.S., Rudik A.V., Filimonov D.A., Poroikov V.V. (2023) HGMMX: Host Gut Microbiota Metabolism Xenobiotics Database, Journal of Chemical Information and Modeling, 63(21), 6463-6468. DOI:10.1021/acs.jcim.3c00837
Kolodnitsky A.S., Ionov N.S., Rudik A.V., Filimonov D.A., Poroikov V.V. (2023) HGMMX: Host Gut Microbiota Metabolism Xenobiotics Database, Journal of Chemical Information and Modeling, 63(21), 6463-6468. DOI:10.1021/acs.jcim.3c00837
![]() Karasev D.A., Sobolev B.N., Lagunin A.A., Filimonov D.A., Poroikov V.V. (2022) The method predicting interaction between protein targets and small-molecular ligands with the wide applicability domain, Computational Biology and Chemistry, 98, 107674. DOI:10.1016/j.compbiolchem.2022.107674
Karasev D.A., Sobolev B.N., Lagunin A.A., Filimonov D.A., Poroikov V.V. (2022) The method predicting interaction between protein targets and small-molecular ligands with the wide applicability domain, Computational Biology and Chemistry, 98, 107674. DOI:10.1016/j.compbiolchem.2022.107674
![]() Tarasova O.A., Rudik A.V., Biziukova N.Yu., Filimonov D.A., Poroikov V.V. (2022) Chemical named entity recognition in the texts of scientific publications using the naïve Bayes classifier approach, Journal of Cheminformatics, 14(1), 55. DOI:10.1186/s13321-022-00633-4
Tarasova O.A., Rudik A.V., Biziukova N.Yu., Filimonov D.A., Poroikov V.V. (2022) Chemical named entity recognition in the texts of scientific publications using the naïve Bayes classifier approach, Journal of Cheminformatics, 14(1), 55. DOI:10.1186/s13321-022-00633-4
![]() Gomazkov O.A. (2021) Neuroproteomics: How a Multitude of Proteins Reflect Brain Functions, Biology Bulletin Reviews, 11(2), 143-153. DOI:10.1134/S2079086421020043
Gomazkov O.A. (2021) Neuroproteomics: How a Multitude of Proteins Reflect Brain Functions, Biology Bulletin Reviews, 11(2), 143-153. DOI:10.1134/S2079086421020043
![]() Dmitriev A.V., Rudik A.V., Karasev D.A., Pogodin P.V., Lagunin A.A., Filimonov D.A., Poroikov V.V. (2021) In Silico Prediction of Drug–Drug Interactions Mediated by Cytochrome P450 Isoforms, Pharmaceutics, 13(4), 538. DOI:10.3390/pharmaceutics13040538
Dmitriev A.V., Rudik A.V., Karasev D.A., Pogodin P.V., Lagunin A.A., Filimonov D.A., Poroikov V.V. (2021) In Silico Prediction of Drug–Drug Interactions Mediated by Cytochrome P450 Isoforms, Pharmaceutics, 13(4), 538. DOI:10.3390/pharmaceutics13040538
![]() Muratov E.N., Amaro R., Andrade C.H., Brown N., Ekins S., Fourches D., Isayev O., Kozakov D., Medina-Franco J., Merz K.M., Oprea T.I., Poroikov V., Schneider G., Todd M.H., Varnek A., Winkler D.A., Zakharov A., Cherkasov A., Tropsha A. (2021) A critical overview of computational approaches employed for COVID-19 drug discovery, Chemical Society Reviews, 50(16), 9121-9151. DOI:10.1039/d0cs01065k
Muratov E.N., Amaro R., Andrade C.H., Brown N., Ekins S., Fourches D., Isayev O., Kozakov D., Medina-Franco J., Merz K.M., Oprea T.I., Poroikov V., Schneider G., Todd M.H., Varnek A., Winkler D.A., Zakharov A., Cherkasov A., Tropsha A. (2021) A critical overview of computational approaches employed for COVID-19 drug discovery, Chemical Society Reviews, 50(16), 9121-9151. DOI:10.1039/d0cs01065k
![]() Poroikov V.V. (2020) Computer-aided drug design: from discovery of novel pharmaceutical agents to systems pharmacology, Biomeditsinskaya Khimiya, 66(1), 30-41. DOI:10.18097/PBMC20206601030
Poroikov V.V. (2020) Computer-aided drug design: from discovery of novel pharmaceutical agents to systems pharmacology, Biomeditsinskaya Khimiya, 66(1), 30-41. DOI:10.18097/PBMC20206601030
![]() Muratov E.N., Bajorath J., Sheridan R.P., Tetko I.V., Filimonov D., Poroikov V., Oprea T.I., Baskin I.I., Varnek A., Roitberg A., Isayev O., Curtarolo S., Fourches D., Cohen Y., Aspuru-Guzik A., Winkler D.A., Agrafiotis D., Cherkasov A., Tropsha A. (2020) QSAR without borders, Chemical Society Reviews, 49, 3525-3564. DOI:10.1039/d0cs00098a
Muratov E.N., Bajorath J., Sheridan R.P., Tetko I.V., Filimonov D., Poroikov V., Oprea T.I., Baskin I.I., Varnek A., Roitberg A., Isayev O., Curtarolo S., Fourches D., Cohen Y., Aspuru-Guzik A., Winkler D.A., Agrafiotis D., Cherkasov A., Tropsha A. (2020) QSAR without borders, Chemical Society Reviews, 49, 3525-3564. DOI:10.1039/d0cs00098a
![]() Druzhilovskiy D.S., Stolbov L.A., Savosina P.I., Pogodin P.V., Filimonov D.A., Veselovsky A.V., Stefanisko K., Tarasova N.I., Nicklaus M.C., Poroikov V.V. (2020) Computational Approaches to Identify a Hidden Pharmacological Potential in Large Chemical Libraries, Supercomputing Frontiers and Innovations, 7(3), 57-76. DOI:10.14529/jsfi200306
Druzhilovskiy D.S., Stolbov L.A., Savosina P.I., Pogodin P.V., Filimonov D.A., Veselovsky A.V., Stefanisko K., Tarasova N.I., Nicklaus M.C., Poroikov V.V. (2020) Computational Approaches to Identify a Hidden Pharmacological Potential in Large Chemical Libraries, Supercomputing Frontiers and Innovations, 7(3), 57-76. DOI:10.14529/jsfi200306
![]() Lloyd K., Papoutsopoulou S., Smith E., Stegmaier P., Bergey F., Morris L., Kittner M., England H., Spiller D., White M.H.R., Duckworth C.A., Campbell B.J., Poroikov V., Martins Dos Santos V.A.P., Kel A., Muller W., Pritchard D.M., Probert C., Burkitt M.D. (2020) Using systems medicine to identify a therapeutic agent with potential for repurposing in inflammatory bowel disease, Disease Models and Mechanisms, 2020(13), dmm044040. DOI:10.1242/dmm.044040
Lloyd K., Papoutsopoulou S., Smith E., Stegmaier P., Bergey F., Morris L., Kittner M., England H., Spiller D., White M.H.R., Duckworth C.A., Campbell B.J., Poroikov V., Martins Dos Santos V.A.P., Kel A., Muller W., Pritchard D.M., Probert C., Burkitt M.D. (2020) Using systems medicine to identify a therapeutic agent with potential for repurposing in inflammatory bowel disease, Disease Models and Mechanisms, 2020(13), dmm044040. DOI:10.1242/dmm.044040
![]() Ivanov S.M., Lagunin A.A., Rudik A.V., Filimonov D.A., Poroikov V.V. (2018) ADVERPred – web service for prediction of adverse effects of drugs, Journal of Chemical Information and Modeling, 58(1), 8-11. DOI:10.1021/acs.jcim.7b00568
Ivanov S.M., Lagunin A.A., Rudik A.V., Filimonov D.A., Poroikov V.V. (2018) ADVERPred – web service for prediction of adverse effects of drugs, Journal of Chemical Information and Modeling, 58(1), 8-11. DOI:10.1021/acs.jcim.7b00568
![]() Filimonov D.A., Druzhilovskiy D.S., Lagunin A.A., Gloriozova T.A., Rudik A.V., Dmitriev A.V., Pogodin P.V., Poroikov V.V. (2018) Computer-aided prediction of biological activity spectra for chemical compounds: opportunities and limitations, Biomedical Chemistry: Research and Methods, 1(1), e00004. DOI:10.18097/BMCRM00004
Filimonov D.A., Druzhilovskiy D.S., Lagunin A.A., Gloriozova T.A., Rudik A.V., Dmitriev A.V., Pogodin P.V., Poroikov V.V. (2018) Computer-aided prediction of biological activity spectra for chemical compounds: opportunities and limitations, Biomedical Chemistry: Research and Methods, 1(1), e00004. DOI:10.18097/BMCRM00004
![]() Ivanov S.M., Lagunin A.A., Poroikov V.V. (2016) In silico assessment of adverse drug reactions and associated mechanisms, Drug Discovery Today, 21(1), 58-71. DOI:10.1016/j.drudis.2015.07.018
Ivanov S.M., Lagunin A.A., Poroikov V.V. (2016) In silico assessment of adverse drug reactions and associated mechanisms, Drug Discovery Today, 21(1), 58-71. DOI:10.1016/j.drudis.2015.07.018
![]() Gomazkov O.A. (2015) Cortexin. Molecular mechanisms and targets of neuroprotective activity, Neuroscience and Behavioral Physiology, 115(8), 99-104. DOI:10.17116/jnevro20151158199-104
Gomazkov O.A. (2015) Cortexin. Molecular mechanisms and targets of neuroprotective activity, Neuroscience and Behavioral Physiology, 115(8), 99-104. DOI:10.17116/jnevro20151158199-104
![]() Lagunin A.A., Goel R.K., Gawande D.Y. Pahwa P., Gloriozova T.A., Dmitriev A.V., Ivanov S.M., Rudik A.V., Konova V.I., Pogodin P.V., Druzhilovsky D.S., Poroikov V.V. (2014) Chemo- and bioinformatics resources for in silico drug discovery from medicinal plants beyond their traditional use: a critical review, Natural Product Reports, 21(11), 1585-1611. DOI:10.1039/C4NP00068D
Lagunin A.A., Goel R.K., Gawande D.Y. Pahwa P., Gloriozova T.A., Dmitriev A.V., Ivanov S.M., Rudik A.V., Konova V.I., Pogodin P.V., Druzhilovsky D.S., Poroikov V.V. (2014) Chemo- and bioinformatics resources for in silico drug discovery from medicinal plants beyond their traditional use: a critical review, Natural Product Reports, 21(11), 1585-1611. DOI:10.1039/C4NP00068D
![]() Gomazkov O.A. (2012) Neurotrophins: The Therapeutic Potential and Concept of Minipeptides, Neurochemical Journal, 6(3), 163-172. DOI:10.1134/S1819712412030075
Gomazkov O.A. (2012) Neurotrophins: The Therapeutic Potential and Concept of Minipeptides, Neurochemical Journal, 6(3), 163-172. DOI:10.1134/S1819712412030075
![]() Filimonov D.A., Zakharov A.V., Lagunin A.A., Poroikov V.V. (2009) QNA based “Star Track” QSAR approach, SAR and QSAR in Environmental Research, 20(7-8), 679-709. DOI:10.1080/10629360903438370
Filimonov D.A., Zakharov A.V., Lagunin A.A., Poroikov V.V. (2009) QNA based “Star Track” QSAR approach, SAR and QSAR in Environmental Research, 20(7-8), 679-709. DOI:10.1080/10629360903438370
![]() Zotchev S.B., Stepanchikova A.V., Sergeyko A.P., Sobolev B.N., Filimonov D.A., Poroikov V.V. (2006) Rational design of macrolides by virtual screening of combinatorial libraries generated through in silico manipulation of polyketide synthases, Journal of Medicinal Chemistry, 49(6), 2077-2087. DOI:10.1021/jm051035i
Zotchev S.B., Stepanchikova A.V., Sergeyko A.P., Sobolev B.N., Filimonov D.A., Poroikov V.V. (2006) Rational design of macrolides by virtual screening of combinatorial libraries generated through in silico manipulation of polyketide synthases, Journal of Medicinal Chemistry, 49(6), 2077-2087. DOI:10.1021/jm051035i
![]() Geronikaki A., Babaev E., Dearden J., Dehaen W., Filimonov D., Galaeva I., Krajneva V., Lagunin A., Macaev F., Molodavkin G., Poroikov V., Pogrebnoi S., Saloutin V., Stepanchikova A., Stingaci E., Tkach N., Vlad L., Voronina T. (2004) Design, synthesis, computational and biological evaluation of new anxiolytics, Bioorganic & Medicinal Chemistry, 12(24), 6559-6568. DOI:10.1016/j.bmc.2004.09.016
Geronikaki A., Babaev E., Dearden J., Dehaen W., Filimonov D., Galaeva I., Krajneva V., Lagunin A., Macaev F., Molodavkin G., Poroikov V., Pogrebnoi S., Saloutin V., Stepanchikova A., Stingaci E., Tkach N., Vlad L., Voronina T. (2004) Design, synthesis, computational and biological evaluation of new anxiolytics, Bioorganic & Medicinal Chemistry, 12(24), 6559-6568. DOI:10.1016/j.bmc.2004.09.016
![]() Poroikov V.V., Filimonov D.A., Ihlenfeldt W.D., Gloriozova T.A., Lagunin A.A., Borodina Y.V., Stepanchikova A.V., Nicklaus M.C. (2003) PASS biological activity spectrum predictions in the enhanced open NCI database browser, Journal of Chemical information and Computer Sciences, 43(1), 228-236. DOI:10.1021/ci020048r
Poroikov V.V., Filimonov D.A., Ihlenfeldt W.D., Gloriozova T.A., Lagunin A.A., Borodina Y.V., Stepanchikova A.V., Nicklaus M.C. (2003) PASS biological activity spectrum predictions in the enhanced open NCI database browser, Journal of Chemical information and Computer Sciences, 43(1), 228-236. DOI:10.1021/ci020048r
![]() Poroikov V.V., Filimonov D.A., Borodina Y.V., Lagunin A.A., Kos A. (2000) Robustness of biological activity spectra predicting by computer program PASS for noncongeneric sets of chemical compounds, Journal of Chemical information and Computer Sciences, 40, 1349-1355. DOI:10.1021/ci000383k
Poroikov V.V., Filimonov D.A., Borodina Y.V., Lagunin A.A., Kos A. (2000) Robustness of biological activity spectra predicting by computer program PASS for noncongeneric sets of chemical compounds, Journal of Chemical information and Computer Sciences, 40, 1349-1355. DOI:10.1021/ci000383k
![]() Lagunin A., Stepanchikova A., Filimonov D., Poroikov V. (2000) PASS: prediction of activity spectra for biologically active substances, Bioinformatics, 16, 747-748. DOI:10.1093/bioinformatics/16.8.747
Lagunin A., Stepanchikova A., Filimonov D., Poroikov V. (2000) PASS: prediction of activity spectra for biologically active substances, Bioinformatics, 16, 747-748. DOI:10.1093/bioinformatics/16.8.747
![]() Filimonov D., Poroikov V., Borodina Y., Gloriozova T. (1999) Chemical similarity assessment through multilevel neighborhoods of atoms: Definition and comparison with the other descriptors, Journal of Chemical information and Computer Sciences, 39(4), 666-670. DOI:10.1021/ci980335o
Filimonov D., Poroikov V., Borodina Y., Gloriozova T. (1999) Chemical similarity assessment through multilevel neighborhoods of atoms: Definition and comparison with the other descriptors, Journal of Chemical information and Computer Sciences, 39(4), 666-670. DOI:10.1021/ci980335o
Phyto4Health database, © Ionov N.S., Filimonov D.A., Druzhilovskiy D.S., Poroikov V.V. Certificate of the Russian State Patent Agency No. 2023622658 of 14.07.2023.
Sarmath program package, © Stolbov L.A., Filimonov D.A., Poroikov V.V. Certificate of the Russian State Patent Agency No. 2022662930 of 07.07.2022.
PASS SMP program package, © Rudik A.V., Dmitriev A.V., Lagunin A.A., Filimonov D.A. Certificate of the Russian State Patent Agency No. 201663627 of 13.12.2016.
PASS Targets program package, © Pogodin P.V., Lagunin A.A., Filimonov D.A., Poroikov V.V. Certificate of the Russian State Patent Agency No. 2016610846 of 20.01.2016.
PASS CLC Pred program package, © Konova V.I., Pogodin P.V., Lagunin A.A., Filimonov D.A., Poroikov V.V. Certificate of the Russian State Patent Agency No. 2016610382 of 11.01.2016.
Net2Target program package, © Ivanov S.M., Filimonov D.A., Lagunin A.A., Poroikov V.V. Certificate of the Russian State Patent Agency No. 2014660877 of 17.10.2014.
NetFlowEx program package, © Koborova O.N., Filimonov D.A., Kel A.E., Poroikov V.V. Certificate of the Russian State Patent Agency No. 2011617330 of 21.09.2011.
HCVmap Database, Sobolev B.N., Kolesanova E.F., Olenina L.V., Rudik A.V., Poroikov V.V. Certificate of the Russian State Patent Agency No. 2007620132 of 10.05.2007.
BIOGENPHARM program package, © Zotchev A.B., Sobolev B.N., Stepanchikova A.V., Sergeiko A.P., Filimonov D.A., Lagunin A.A., Gloriozova T.A., Poroikov V.V. Certificate of the Russian State Patent Agency No. 2006614395 of 15.02.2007.
BIOGENERATOR program package, © Zotchev A.B., Sobolev B.N., Stepanchikova A.V., Sergeiko A.P., Poroikov V.V. Certificate of the Russian State Patent Agency No. 2006614396 of 15.02.2007.
GUSAR (General Unrestricted Structure-Activity Relationships) program package, © Zakharov A.V., Filimonov D.A., Lagunin A.A., Poroikov V.V., Certificate of the Russian State Patent Agency No. 2006613591 of 16.10.2006.
PharmaExpert program package, © Lagunin A.A., Poroikov V.V., Filimonov D.A., Gloziozova T.A. Certificate of the Russian State Patent Agency No. 2006613590 of 16.10.2006.
PreTox program package, © Filimonov D.A., Poroikov V.V., Gloziozova T.A., Lagunin A.A. Certificate of the Russian State Patent Agency No. 2006613276 of 15.09.2006.
PASS program package, © Filimonov D.A., Poroikov V.V., Gloziozova T.A., Lagunin A.A. Certificate of the Russian State Patent Agency No. 2006613275 of 15.09.2006.
METAPREDICT (Prediction of Biotransformations) program package, © Poroikov V.V., Filimonov D.A., Borodina Yu.V., Rudik A.V. Certificate of the Russian State Patent Agency No. 2004610666 of 12.03.2004.
- Alexander Kel, Galina Selivanova, Audrey Shchekotikhin, Galina Buravchenko, Vladimir Koutcherov, Vladimir Poroikov, Roman Zoubarev, Elias Arnér.. Quinoxaline thioredoxin reductase inhibitors. Application filed by Genexplain Gmbh 2022-12-23. Publication of WO2023118593A1 2023-06-29.
- A.E. Shchekotikhin, G.N. Selivanova, V.V. Poroikov, A.V. Zakharov, A.E. Kel, V.G. Kutcherov. Method for treating a tumor disease and method for selectively inhibiting tumor cell growth using a quinoxaline-1,4-dioxide derivative. Application number PCT / RU2014 / 000310, publication number WO2015167350 A1, priority date Apr 28, 2014, publication date Nov 5, 2015.
- A.E. Shchekotikhin, G.N. Selivanova, V.V. Poroikov, A.V. Zakharov, A.E. Kel, V.G. Kutcherov. A method for treating a tumor disease and a method for selectively inhibiting the growth of tumor cells using a quinoxaline-1,4-dioxide derivative. Eurasian patent No. 031473. Date of filing: April 28, 2014. Date of issue of the patent: January 21, of filing: April 28, 2014. Date of issue of the patent: January 21, 2019.
Dr.Sci.
- Lagunin A.A. “Computer-Aided Estimation of Pleiotropic Action of Pharmacological Substances”.
- Kolpakov F.A. “Computer Modeling of Biological Systems and Analysis of Biomedical Data”.
Ph.D.
- Akimov D.V. “Computer-Aided Search for New HIV-1 Integrase Inhibitors”.
- Borodina Yu.V. “Computer Analysis of Similarity between Pharmaceutical Substances and Endogenous Bioregulators and Their Synthetic Analogues”.
- Dmitriev A.V. “Computer-Aided Prediction of Xenobiotics’ Interaction with Human Cytochromes P450”.
- Druzhilovskiy D.S. “Search and Optimization of New HIV-1 Integrase Inhibitors on the Basis of Computer-Aided Predictions”.
- Dubovskaja (Konova) V.I. «Computer-Aided Finding of Chemicals with Cytotoxic Action on Breast Cancer Cell Lines».
- Eremeevskaya (Koborova) O.N. “Search for Pharmacological Targets for Breast Cancer Therapy on the Basis of Computational Modeling of Cell Cycle”.
- Karasev D.A. “Development of a Proteochemometrics Method for Predicting Protein-Ligand Interactions Based on Their Local Similarity”.
- Lagunin A.A. “Search for New Biologically Active Compounds on the Basis of Computer Analysis of Structure-Mechanisms-Effects Relationships”.
- Linde D.M. “Intermolecular Recognition in Globular Proteins from Brookhaven’s Data Bank”.
- Pogodin P.V. “Computer-Aided Estimation of Interaction of Drug-Like Organic Compounds with Human Kinome”.
- Romero Reyes I.V. “Estimating affinity of protein-ligand complexes using neural networks”.
- Rudik A.V. “Computer Prediction of Xenobiotic’s Biotransformations”.
- Sergeyko A.V. “Computer-Aided Design of Enzymatic Systems Providing Biosynthesis of Macrolides with the Required Properties”.
- Stolbov L.A. “Development of Approaches to Virtual Screening of Antiviral Compounds Taking into Account the Heterogeneity of Information”.
- Tarasova (Filz) O.A. “Design of compounds with the required biological activity by a combination of functionally-important fragments”.
- Zakharov A.V. “Prediction of qualitative properties of organic compounds on the basis of descriptors of atomic neighborhoods”.
Diploma Works
- Akimov D.V., N.I. Pirogov Russian National Research Medical University. “Computer Search for HIV-1 Integrase Inhibitors”.
- Aleksandrov K.E., N.I. Pirogov Russian National Research Medical University. “Information Resource on Functional Mapping of Hepatitis C viral proteins”.
- Bezhentsev V.M., Higher School of Economics. “Search for Novel Inhibitors of HIV Integrase Using Methods of Structural Bioinformatics”.
- Biziukova N.Yu., Pirogov Russian National Research Medical University. “Development of an algorithm for extracting information about chemical compounds and human proteins from publications”.
- Veselova D.A., Pirogov Russian National Research Medical University. “Analysis of three-dimensional patterns of enzymatic specificity of protein phosphorylation using the developed software tool”. RSM
- Dmitriev A.V.,N.I. Pirogov Russian National Research Medical University. “Computer Prediction of Chemical Compounds Biotransformations by CYP3A4”.
- Druzhilovskiy D.S., N.I. Pirogov Russian National Research Medical University. “Computer-Aided Prediction of HIV-1 Integrase Inhibitors”.
- Filz (Tarasova) O.A., N.I. Pirogov Russian National Research Medical University. “Computer-Aided Design of Organic Compounds with the Required Properties from the Libraries of Structural Fragments”.
- Fomenko A.E., N.I. Pirogov Russian National Research Medical University. “Analysis of Structure-Function Relationships in Serine Proteases by Bioinformatics Methods”.
- Ionov N.S., N.I. Pirogov Russian National Research Medical University., “Computer-Aided Analysis of Application of Natural Compounds for Therapy of Metabolic Diseases”.
- Ivanov S.M., N.I. Pirogov Russian National Research Medical University. “Computational Search for Molecular Targets Associated with Adverse Effects of Non-Steroid Anti-Inflammatory Drugs”.
- Kabieva Sh.Sh. , N.I. Pirogov Russian National Research Medical University. “Sechenov University. Analysis of Signaling Pathways in the Progressing of HIV Infection Using Bioinformatics Methods”.
- Karasev D.A., N.I. Pirogov Russian National Research Medical University. “Analysis of Amino Acid Sequences Responsible for Specificity of Protein Substrates to Protein Kinases MAPK and CDK”.
- Koborova (Eremeyevskaya) O.N., M.V. Lomonosov Moscow State University. “Development of Method for Estimating Antineoplastic Targets Promising for Breast Cancer Therapy”.
- Konova (Dubovskaja) V.I., N.I. Pirogov Russian National Research Medical University. “Search for Promising Pharmacological Targets for Therapy of Alzheimer’s Disease”.
- Korotkevich E.I., N.I. Pirogov Russian National Research Medical University. “Development of a computer method for predicting the metabolic stability of xenobiotics”.
- Lagunin A.A., N.I. Pirogov Russian National Research Medical University. “Expert System on Relationships between Pharmacological Effects and Mechanisms of Actions in Biologically Active Compounds”.
- Levchenko M.E.,N.I. Pirogov Russian National Research Medical University. “Receptors of Excitatory Amino Acids: Computer Search for New Ligands and Analysis of Mechanisms”.
- Martynova N.B., N.I. Pirogov Russian National Research Medical University. “Creation of Virtual Combinatory Libraries of Oligopeptides and Peptidomimetics for Search of New Lead Compounds”.
- Murtazalieva Kh.A., N.I. Pirogov Russian National Research Medical University. “Computer-Aided Finding and Experimental Validation of Pharmaceutical Agents Acting on MDA-MB-231 Cell Line”.
- Paremskaya A.Yu. , N.I. Pirogov Russian National Research Medical University. “Computational Prediction of HIV Drug Resistance”.
- Peredkova L.S., N.I. Pirogov Russian National Research Medical University. “Analysis of Differential Gene Expression of Blood Cells from HIV Patients”.
- Pogodin P.V., N.I. Pirogov Russian National Research Medical University. “Computer-Aided Prediction of Low Molecular Weight Organic Compounds Interaction with Protein Targets Based On PASS Software and ChEMBL Database”.
- Potapov V.Yu., N.I. Pirogov Russian National Research Medical University. “About the Role of Electrostatic Interaction in Formation of Protein-Protein Complexes”.
- Sadym (Rudik) A.V., Moscow Physical-Engineering Institute. “Development of Internet System for Prediction of Biological Activity Spectra”.
- Saulin P.S., N.I. Pirogov Russian National Research Medical University. “Peculiarities of Amino Acid Composition of T-epitopes”.
- Savosina P.I., N.I. Pirogov Russian National Research Medical University, "Development of Methods for “Big Data” Analysis to Discover New Anti-HIV Agents".
- Semin M.I., N.I. Pirogov Russian National Research Medical University. “Computational Approach to Estimation of Drug-Drug Interactions Causing the Hepatotoxicity.
- Serezhenkina I.V., N.I. Pirogov Russian National Research Medical University. “Functional and Antigenic Mapping of the Amino Acid Sequences of Structural Proteins of Flaviviridae Viruses”.
- Sergeyko A.V., N.I. Pirogov Russian National Research Medical University. “Computation of Macrolide’s Chemical Structure on the Basis of Domain Structure of Protein Synthesize Complex”.
- Sukhachev V.S., MIREA – RTU, "Computer-Aided Prediction of Arrhythmogenic Action of Pharmaceutical Agents Based on Analysis of Molecular Networks” (Undergraduate).
- Veselova D.A., N.I. Pirogov Russian National Research Medical University. “Analysis of 3D Patterns of Enzyme Specificity of Protein Phosphorylation with the Developed Software Tools”.
- Urusova A.F., N.I. Pirogov Russian National Research Medical University. “Development of an algorithm for selection of data for creating quantitative structure-activity relationship models using HIV-1 reverse transcriptase as a case study”.
- Yazykova E.I., MIREA – RTU. “Assessment of the Role of Particular Substructures of Drug-Like Compounds in the Manifestation of Biological Activity in Multiple Trials in Vitro”.
- Zakharov A.V., N.I. Pirogov Russian National Research Medical University. “Quantitative Structure-Activity Relationships of Cyclin-Depended Kinase 1 Inhibitors”.